This package facilitates the reading, processing and visualization of N2O isotope data including drift correction, 17O correction and standards calibration.
Installation
As of version 0.4.0, the package is based on the functionality provided by the isoreader and isoprocessor packages to load IRMS data directly from their raw data files and process it. All can be installed from CRAn or directly from GitHub using the devtools package (see troubleshooting section below in case of issues with devtools installs).
install.packages("devtools") install.packages("isoreader") devtools::install_github("isoverse/isoprocessor") devtools::install_github("sebkopf/isorunN2O", build_vignettes = TRUE)
For the earlier version that used the now deprecated isoread instead, run devtools::install_github("sebkopf/isorunN2O", ref = "v0.3.0"). This version unfortunately is no longer supported on newer R versions.
How to use?
The package includes a tutorial that introduces most functionality and provides examples of how it works and can be used. After the isorunN2O package is installed, the newest version of this tutorial can always be loaded up as a vignette directly in R by calling vignette("N2O_data_reduction_tutorial") in the command line. The RMarkdown file underlying the vignette is also available directly in this repository including the resulting HTML output.
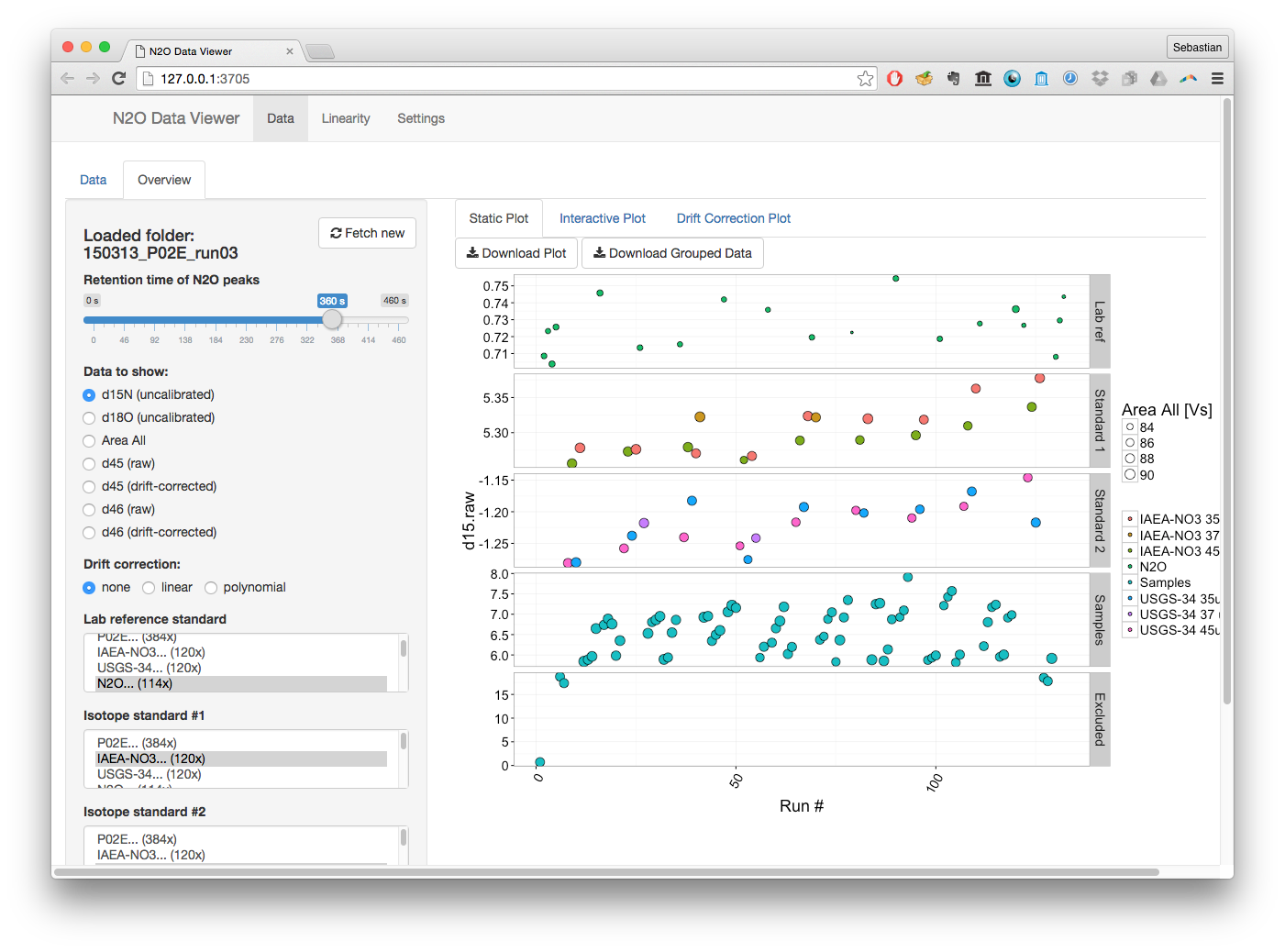
Additional functionality: N2O Data Viewer
Earlier versions of this package (up to 0.3.0) also include a browser based application for interactive N2O run monitoring. It can be launched with the function run_data_viewer(), additional information is available here.
For newer versions of this package (>= 0.4.0), this functionality is mostly covered by the much broader graphical user interface of the isoviewer package although it does not yet recreate all of the original functionality of the N2O data viewer. We ask for patience while isoviewer is still in development, it will eventually provide all the original functionality and much more.
Troubleshooting
On Windows, the above install_github calls can fail because of the requirement to create zip archieves during some vignette creation steps. If this is the case, install Rtools, restart R and RStudio and try to run the installation again.
If you run into any trouble installing the packages that is not resolved easily, try installing them again without the vignettes, which are sometimes the sole culprits, i.e. devtools::install_github(...., build_vignettes = FALSE), which should provide the fully functional package, just without the vignette tutorial (which you can still check out online as mentioned in the How to use? section).
Issues with automatic dependency installation
The latest version of the devtools package on CRAN (version 1.12.0) has a bug that causes issues with the installation of package dependencies, especially on Windows, which usually manifest as errors during the installation of GitHub packages stating some dependency (e.g. stringi or dplyr or htmltools, etc. package missing). Please check this troubleshooting Gist for details if you encounter this error.

